Seattle Children’s Research Institute’s Genomics and Spatial Biology Collaborative Laboratory provides access and specialized support for spatial biology research including transcriptomics, proteomics, genomics and related methodologies.
Spatial Biology Equipment and Services
The Genomics and Spatial Biology CoLab facilitates the measurement of proteins and transcripts in tissue sections using the NanoString GeoMx Digital Spatial Profiler (DSP) platform. Spatially resolved transcriptomics was named Nature Methods’ 2020 Method of the Year. Unlike bulk cell or tissue experiments, digital spatial profiling preserves spatial information and allows transcriptomic and proteomic quantification within defined regions of fixed tissue. This is particularly suited for the characterization of rare cell types and specific microenvironments within tissue. For more info, see the original paper describing GeoMx DSP technology and a recent publication from Seattle Children's Research Institute researchers applying DSP to liver-stage malaria.
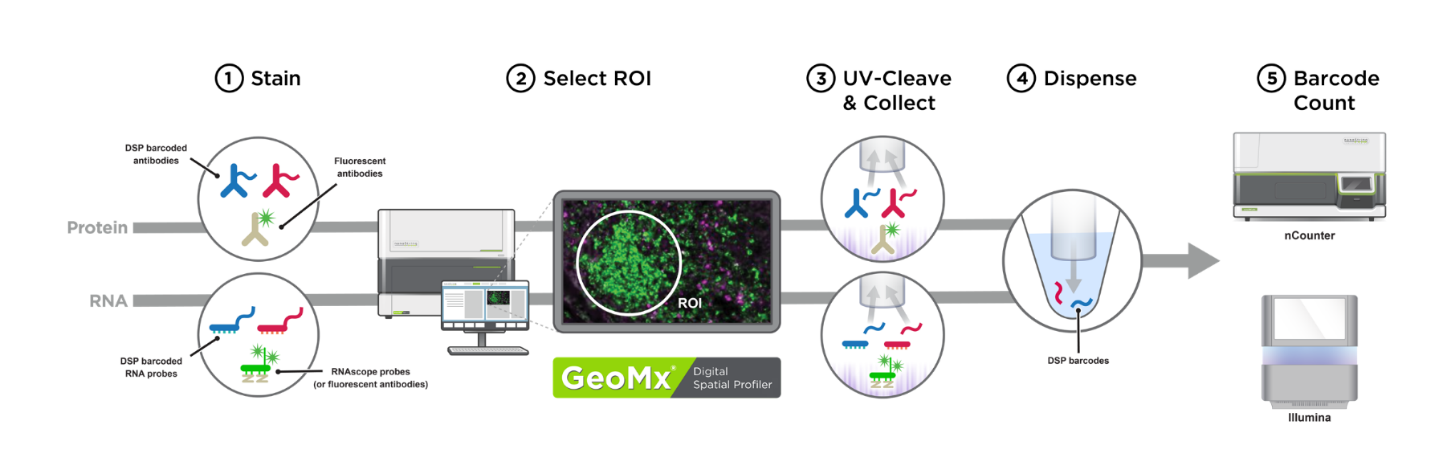
Image source: NeoGeomics Laboratories
The CosMx Spatial Molecular Imager (SMI) is a single-cell imaging platform that enables high-plex in situ analysis for spatial multiomics applications with formalin fixed paraffin embedded (FFPE) and flash frozen (FF) tissue samples at both cellular and subcellular resolution. This instrument provides flexible and robust spatial single-cell imaging for applications of oncology, neuroscience, immunology, infectious disease and developmental biology research for insights including:
- Cell atlasing and characterization
- Cell to cell interaction
- Spatial biomarkers
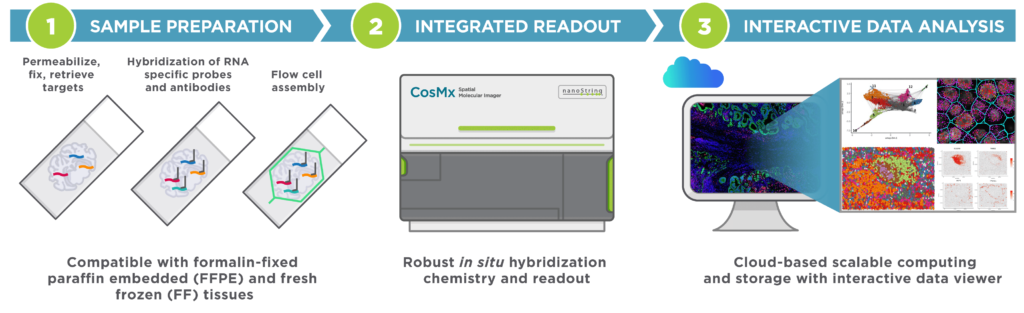
Image source: NanoString
After collection, samples from the GeoMx may be prepped for quantification on the nCounter. The NanoString nCounter SPRINT Profiler combines the nCounter Pro Prep Station and Digital Analyzer into one instrument to allow for the digital examination of multiple pathways in a single tube. Output files include gene names and associated count numbers.

Image source: NanoString
Genomics Equipment and Services
The PacBio Revio is a state of the art long read sequencing system that is particularly useful for genome assembly, structural variant detection, and the study of complex genomic regions. Long read “HiFi” sequencing capabilities provided by the Revio will support investigators who have needs in:
- De novo genome assembly and structural variation analysis
- Transcriptomics (including full-length RNA transcripts)
- Epigenomics (DNA methylation patterns at high resolution/accuracy)
- Functional genomics (identifying function of long non-coding RNAs)
- Microbial genomics
- Cancer genomics (tumor biology/evolution, RNA fusions)
Use of the Revio will be aided by the integration of an Agilent Femto Pulse and Diagenode Megaruptor to facilitate sample QC and shearing for optimal molecular weight.

Image source: PacBio
The Illumina NextSeq 2000 is a desktop sequencer with multiomics applications including whole-exome, spatial transcriptomics and single-cell sequencing. After collection on the GeoMx, samples may be prepped for sequencing on the NextSeq 2000.

Image source: Illumina
The Chromium X combines high throughput single cell gene expression and high throughput immune profiling to perform any 10x Genomics single cell assay at the desired scale, from hundreds to hundreds of thousands of cells per run. New high throughput assays include those for Single Cell Immune Profiling and Single Cell Gene Expression.

Image source: 10x Genomics
The 4200 TapeStation is a high throughput automated electrophoresis platform for nucleic acid sample quality control. The system provides nucleic acid analyses, a wide range of RNA and DNA ScreenTape applications, full sample scalability from 1 to 96 samples, reduced sample consumption and no risk of cross contamination.

Image source: Agilent
The Mission Bio Tapestri uses microfluidic droplet technology to deliver a high throughput single-cell genomics workflow for targeted DNA sequencing. The Tapestri integrates SNV, CNV, and protein data from the same cell.
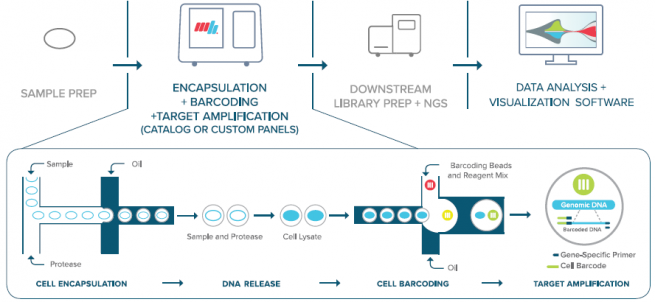
Image source: Mission Bio
Request Services
Genomics and Spatial Biology CoLab services are available for internal and external users. To request services provided by the core, please visit the Genomics and Spatial Biology CoLab’s iLab page.
For information on how to access iLab, refer to this guide.
Genomics and Spatial Biology Rates
To view internal rates for genomics and spatial biology services, please visit this page.
Acknowledging Core Contributions
When citing the Genomics and Spatial Biology CoLab, we recommend the following language: Seattle Children's Research Institute Genomics and Spatial Biology Collaborative Laboratory, RRID:SCR_025490.
If you need more information for your grant, please contact the Genomics and Spatial Biology CoLab staff.
Access and Contact Information
Address for spatial biology and genomics services
Genomics and Spatial Biology CoLab
Seattle Children's Research Institute
1900 Ninth Ave., 7th Floor
Seattle, WA 98101
- General: GenomicsAndSpatialBiology@seattlechildrens.org
- Rebecca Martin, PhD: rebecca.martin@seattlechildrens.org
- Nasaro Kim, MS, MB (ASCP): nasaro.kim@seattlechildrens.org
