Proteomic and Genetic Insight Into Axon Termination, Synapse Maintenance and Autophagy
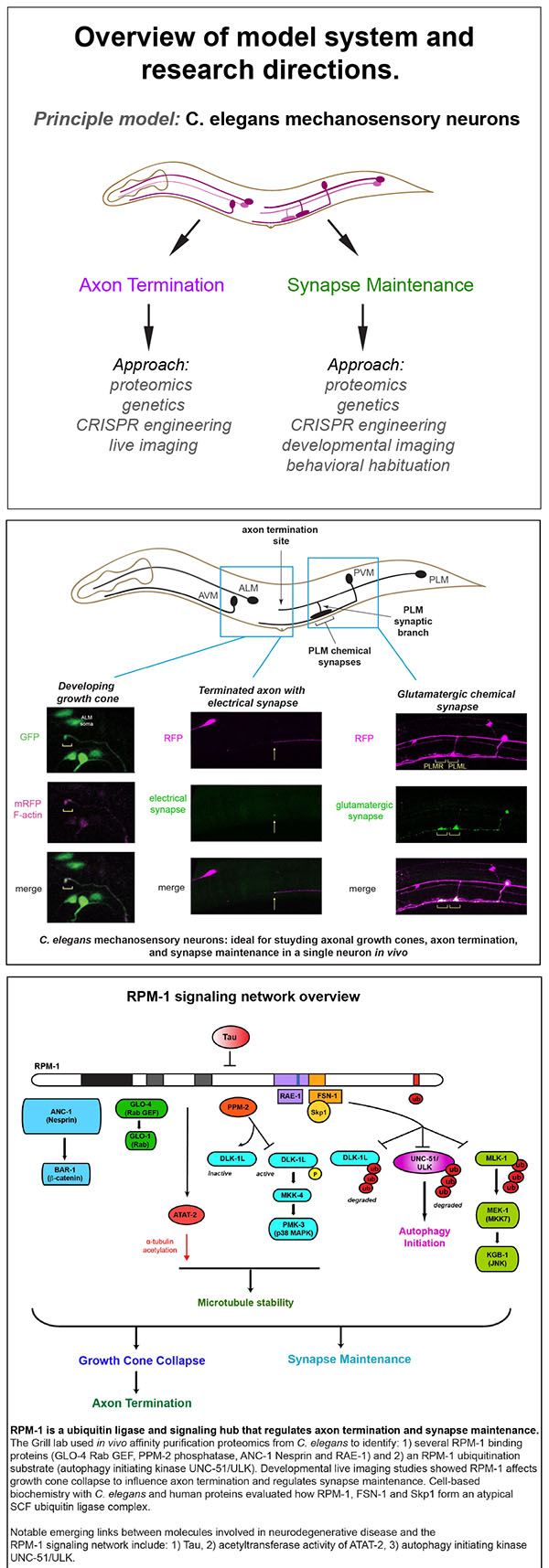
The Grill Lab has made important contributions to understanding signaling networks that regulate axon termination, synapse maintenance and behavioral plasticity. We rely upon C. elegans mechanosensory neurons as our principal in vivo model system. Importantly, much of our progress stems from my group’s status as a leader in C. elegans neural proteomics, which we often use as a starting point for discovery science. Recently, affinity purification proteomics laid the foundation for our exciting discovery that ubiquitin ligase activity inhibits autophagy in the nervous system.
We are leading efforts to understand a conserved signaling network required for both axon termination and synapse maintenance, two fundamental events in nervous system development and stability. At the heart of this network is Regulator of Presynaptic Morphology 1 (RPM-1), a ubiquitin ligase and signaling hub.
Our proteomics work established RPM-1 as the first example of a ubiquitin ligase that acts as a signaling hub in the nervous system. Live imaging and developmental time course approaches have allowed us to study the cellular processes that RPM-1 regulates. Behavioral outcomes indicate that perturbing the RPM-1 signaling network affects behavioral plasticity to repeated touch due to effects on glutamatergic chemical synapses.
In recent work, we used C. elegans proteomics and a ubiquitin ligase biochemical “trap” to identify RPM-1 ubiquitination substrates. This unbiased approach identified the autophagy initiating kinase UNC-51/ULK as an RPM-1 substrate. Genetic, transgenic and CRISPR editing/engineering approaches demonstrated that RPM-1 ubiquitin ligase activity restrains initiation of autophagy in mechanosensory neurons and broadly in the nervous system of C. elegans.
This is just the beginning of what will be a leap forward in neural proteomics in the coming years!
