Publication Q&A: A Systems-Level Gene Regulatory Network Model for Plasmodium Falciparum
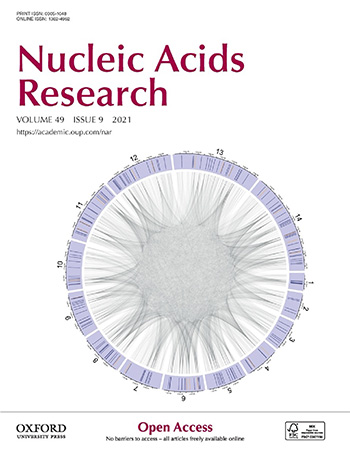 Image source: Nucleic Acids Research, Oxford Univeristy Press
Image source: Nucleic Acids Research, Oxford Univeristy PressMay 2021 – Maxwell Neal, Ling Wei, Alexis Kaushansky and John Aitchison from the Center for Global Infectious Disease Research at Seattle Children’s share insights from a recent publication in Nucleic Acids Research.
This research represents a collaboration between the Aitchison and Kaushansky labs at Seattle Children’s, as well as the Baliga Lab at the Institute for Systems Biology.
An image generated for the paper was used for the cover of the Nucleic Acids Research (Volume 49, Issue 9, 21 May 2021) in which the paper appears. The image is a visualization of the gene regulatory model we generated. The caption for the cover art reads:
Visualization of model-generated gene regulatory network for the malaria parasite Plasmodium falciparum genome. Ring segments represent chromosomes. Orange and blue lines inside segments indicate genomic locations of ApiAP2 and non-ApiAP2 transcriptional regulators, respectively. Edges indicate regulator-target interactions positioned according to the location of their genes.
A systems-level gene regulatory network model for Plasmodium falciparum
Maxwell L Neal, Ling Wei, Eliza Peterson, Mario L Arrieta-Ortiz, Samuel A Danziger, Nitin S Baliga, Alexis Kaushansky, John D Aitchison
Read this publication in Nucleic Acids Research.
What are the significant findings in this paper?
This paper presents the first comprehensive model of how gene expression is controlled in the deadliest malaria parasite Plasmodium falciparum. This model allows researchers to investigate how the parasite regulates gene expression and to identify regulatory genes that are most critical during the symptomatic stage of malaria infection. For example, the model predicts the existence of several “hub” genes that each influence the expression of many gene targets. Several of these hubs are known to be critical for parasite infections, but the hub predicted to have the highest level of influence is one that is not well-studied. The model therefore provides signposts for where to focus future research. The model also provides a guide for identifying potential malaria drug targets. For example, disrupting the operation of the hub genes pharmacologically could potentially reduce malaria symptoms.
Developing the model also presented an opportunity to investigate which parts of the parasite’s molecular machinery contribute to artemisinin resistance. Artemisinin is widely used to treat malaria, but over the past two decades, parasites in some countries have become more able to tolerate artemisinin treatment. Surprisingly, our analysis found that artemisinin-resistant parasites had more variability in gene expression patterns, and this variability was associated with many different parts of the parasite’s regulatory machinery. This was surprising because it did not fit the traditional thinking about how resistance originates in microbes. It is commonly thought that resistance arises because mutations on one or a small number of genes change the parasite’s molecular processes in a way that gives them more tolerance to a drug. However, we found that the major difference between drug-resistant and drug-sensitive parasites was that populations of parasites that led to resistance were more diverse in their gene expression patterns. In other words, parasite infections with a higher diversity are more able to resist drugs. This led us to conclude that artemisinin resistance is best understood as the result of parasite infections that have more diversity in their population: It may be that such populations are better equipped to evade the human immune system – like a bed hedging strategy – “diversify so that some of us can survive” and establish infection sites. This discovery, which links drug resistance to the parasite’s strategies for escaping elimination by the human immune system, is critical for understanding the origins of malaria parasite resistance and for determining approaches for combatting the growing challenge of artemisinin resistance.
What are the broad implications of this research?
The model we generated indicates which genes may be critical “hub” controllers used by the parasite during infection, and these controllers are potential drug target candidates: disrupting their function pharmacologically would likely help stop an infection. More generally, the model provides a large number of predictions about how genes are controlled in the parasite. Therefore, it can be used by researchers as a comprehensive roadmap for understanding the way the organism operates at a molecular level, which can lead to new approaches for treating malaria.
Our discovery linking gene expression diversity with artemisinin resistance provides a new, valuable framework for understanding how resistance has emerged and where to focus future research aimed at targeting resistant parasite populations. It may help researchers better predict how well an individual’s infection will respond to artemisinin treatment and may help in global surveillance efforts that aim to predict the emergence of resistant populations.
What are the next steps and long-term goals for this research?
Members of the Aitchison and Kaushansky labs are currently writing a grant proposal that focuses on identifying the genes and molecular mechanisms that make a parasite population more diverse and better able to tolerate drug pressure.
The labs also plan to investigate several of the “hub” regulator genes predicted by the model to better understand their role in malaria infections.
A more long-term goal is to apply the same modeling approach used in this research to determine gene regulatory patterns in other phases of the parasite life cycle. It will be interesting to see how the global gene regulatory network used by the parasite during its mosquito stage differs from the network used during human blood-stage infection.
